RESEARCH
APOL1 and Lupus
Collapsing Lupus Glomerulopathy
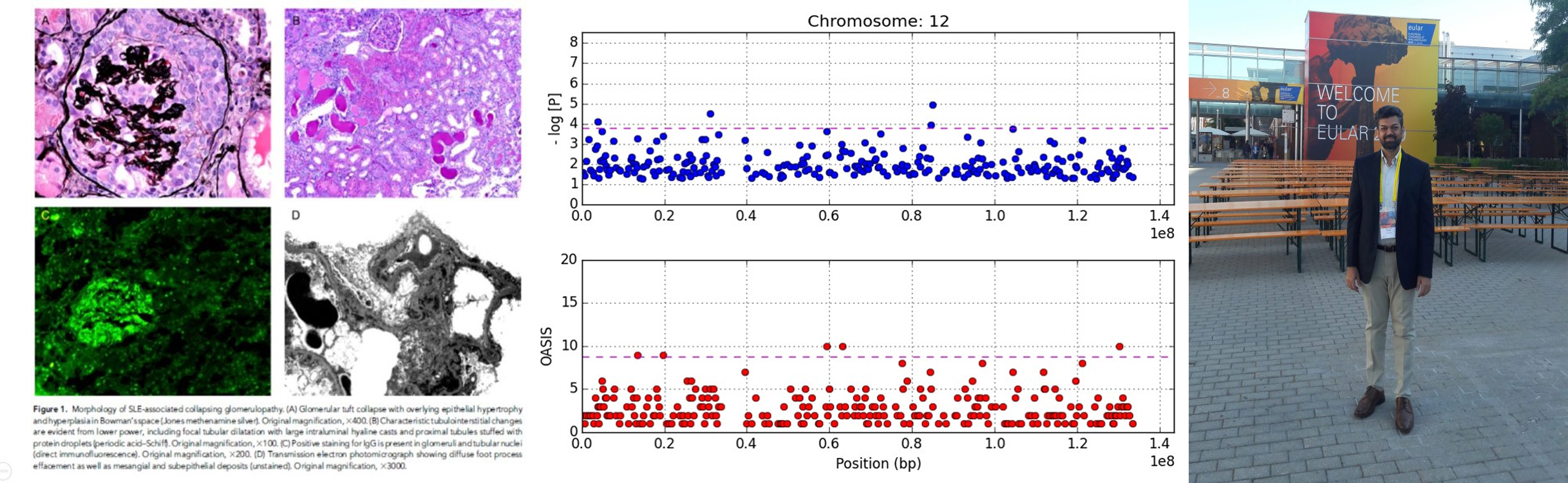
Systemic lupus erythematosus (SLE) is a clinically elusive disease with myriad features. The other challenge in SLE is the relatively low frequency of disease making large-sized sample collection for genetic studies difficult. One of the most definitive finding of SLE is lupus nephritis (LN). In association with Nephropath (presently Arkana Labs) in 2012, Dr. Mohammad Saeed designed a modified case–control genetic association study using tissue biopsy samples. This novel design made possible the study of pathologically well-defined SLE phenotype (LN) in large enough numbers for meaningful analysis. Dr. Marjorie Beggs skillfully extracted the DNA from FFPE samples and tweaked the PCR protocols to allow genotyping with TaqMan assays. Dr. Chris Larsen provided the Pathologic expertise and disease insight and Prof. Patrick Walker (CEO Nephropath) provided the funding, project oversight and the huge number of well-curated pathology samples for the genetic analysis. This unique combination of people and resources led to the discovery of APOL1 alleles conferring over 5x fold risk of developing SLE-associated collapsing glomerulopathy (SLECG). It was the first time significant insight had been gained in this devastating renal disease that lacks treatment so far. Diagnosis is key here because it can save the patient from potential toxic therapies, such as high dose steroids and immunosuppressants that have not been shown to be of benefit in SLECG, while being of immense benefit in SLE and LN. This study was cited as one of “Top 10 Developments in Lupus Nephritis” in 2013. J Am Soc Neph (JASN) 2013APOL1 Histopathology
Morphologic findings in kidney biopsies from patients with chronic kidney disease (CKD) are generally nonspecific and are often labeled ‘hypertensive’ or arteriolar nephrosclerosis. The injury patterns represent final common pathways for multiple forms of renal disease. We showed that there are in fact, specific histopathologic findings that are more frequently associated with APOL1 risk alleles. Solidified and disappearing glomerulosclerosis, mean degrees of interstitial fibrosis, thyroidization-type tubular atrophy and microcystic tubular dilatation were significantly greater in the presence of two APOL1 risk variants. Simultaneously, obsolescent glomerulosclerosis and greater degrees of arteriosclerosis were present significantly more often in those without the APOL1 risk variants. Mod Path 2015APOL1 Clinical Genetic Test
Finally, this effort resulted in the development of the APOL1 clinical test which Arkana Labs now offers as part of its Molecular Pathology service. Hence, the novel tissue-based genetic association study design led to multi-fold benefit encompassing accurate clinical description of disease phenotype (SLECG), discovery of the underlying genetic cause (APOL1 in this case), clinical pathology correlates (specific histopathologic features) as markers of genetic disease as well as a simple blood test that can potentially avoid the invasive kidney biopsy. In short, it was Translational Medicine at its best.PLA2R and Membranous Glomerulonephritis
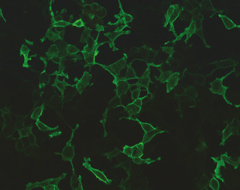
Immunofluorescent staining for PLA2R Antibodies
Membranous Glomerulopathy (MG) MG is a morphologic pattern of glomerulonephritis in which immune complexes deposit along the subepithelial aspect of the glomerular basement membrane. The molecular etiology of MG was recently discovered to be Podocyte Phospholipase A2 receptor (PLA2R) and its genetic susceptibility variant was identified by Dr. Mohammad Saeed and colleagues (Saeed et al. 2014). Using the design by Dr. Mohammad Saeed, of a modified case–control genetic association study on tissue biopsy samples, MG could be studied in large numbers with phenotypic accuracy i.e. in accordance with PLA2R antibody status (positive or negative). It was found that the G-allele of the PLA2R1 variant rs35771982 (His>Asp at postion 300) led to increased antigenicity, perhaps leading to the generation of PLA2R antibodies. This variant was strongly associated with PLA2R antibody positive MG (Saeed et al. 2014). Furthermore, we showed that APOL1 variants in PLA2R+ MG accelerates disease and is a risk factor for collapsing GN (AJKD 2014).
Using the design by Dr. Mohammad Saeed, of a modified case–control genetic association study on tissue biopsy samples, MG could be studied in large numbers with phenotypic accuracy i.e. in accordance with PLA2R antibody status (positive or negative). It was found that the G-allele of the PLA2R1 variant rs35771982 (His>Asp at postion 300) led to increased antigenicity, perhaps leading to the generation of PLA2R antibodies. This variant was strongly associated with PLA2R antibody positive MG (Saeed et al. 2014). Furthermore, we showed that APOL1 variants in PLA2R+ MG accelerates disease and is a risk factor for collapsing GN (AJKD 2014).
PLA2R and Lupus
PLA2R1 gene variants were subsequently shown to be associated with SLE and PLA2R antibodies were detected in Lupus Nephritis at a frequency of 5%. Of the SLE patients with MG, 19% were positive for PLA2R antibodies in serum. The presence of PLA2R antibodies indicated aggressive disease with worse prognosis. These antibodies were present before clinical onset of MG and predict a favorable response to Rituximab.
These findings are a major advance in the study of MG, and has since then led to developments in diagnosis and treatment of the disease. Antibodies against PLA2R are detected in 70-80% of patients with primary MG (Larsen et al. 2014). Previously, diagnosis of autoimmune MG could only be made by kidney biopsy, histological examination or electron microscopy of the kidney tissue. These procedures are invasive, time consuming and costly.PLA2R serum antibody test for clinical diagnosis
Now at ImmunoCure we offer testing of PLA2R antibody on blood samples using the sensitive indirect immunofluorescence (IIFT) method. Post Reference: Saeed M et al. (2014). Genes Immun. PMID: 25187357.DDX11 and Lupus
 Systemic lupus erythematosus (SLE) is a complex disorder manifesting as a syndrome. In SLE, identification of susceptibility genes is of great relevance for understanding specific pathobiological mechanisms and expanding the number of molecular targets for clinical testing and drug discovery.
Genotyping a multitude of single-nucleotide polymorphisms (SNPs) in GWAS leads to multiple testing and proportional signal problems, preventing modest associations from being distinguished from random noise. This is dealt with by multiple testing corrections, which directly result in a high false negativity rate leading to missing heritability, a major cause of disappointment with GWAS. The other approach to deal with the inherent false positivity has been association binning methods such as gene-based testing.
Systemic lupus erythematosus (SLE) is a complex disorder manifesting as a syndrome. In SLE, identification of susceptibility genes is of great relevance for understanding specific pathobiological mechanisms and expanding the number of molecular targets for clinical testing and drug discovery.
Genotyping a multitude of single-nucleotide polymorphisms (SNPs) in GWAS leads to multiple testing and proportional signal problems, preventing modest associations from being distinguished from random noise. This is dealt with by multiple testing corrections, which directly result in a high false negativity rate leading to missing heritability, a major cause of disappointment with GWAS. The other approach to deal with the inherent false positivity has been association binning methods such as gene-based testing.
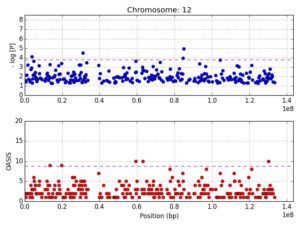
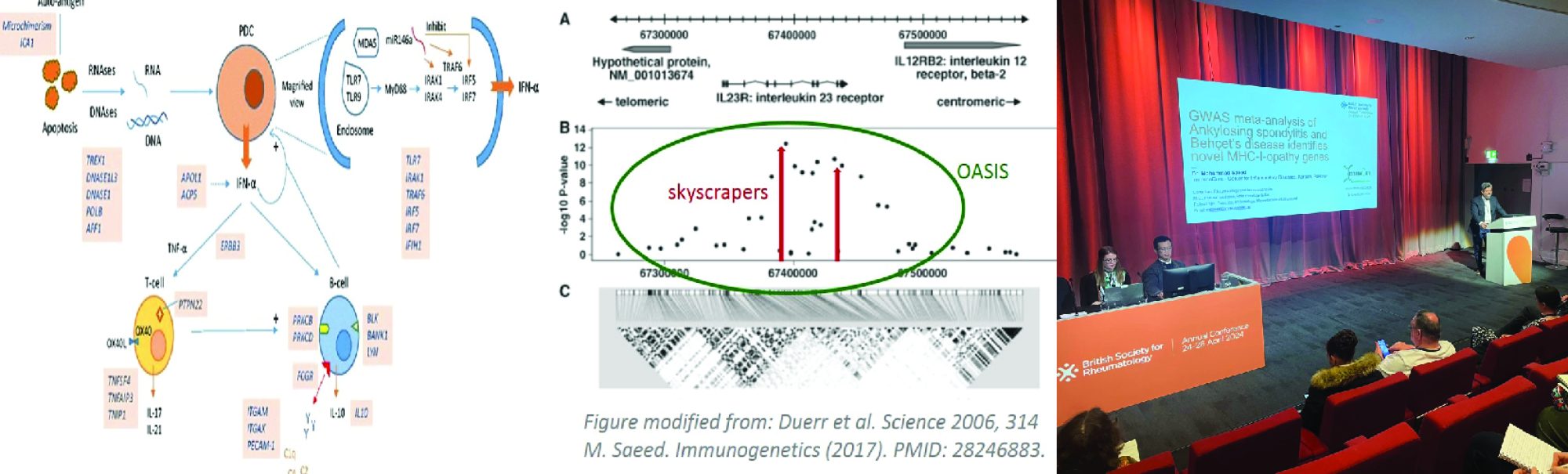
Figure 1 shows the mapping of the 12p11 locus using OASIS analysis of GWAS
Binning methods, by decreasing the number of tests per study, potentially increase power and reduce the multiple-testing burden. Caveats with such approaches include skewing of the association signal due to weighting procedures and to the large extent of LD in historically more recent mutations. OASIS, provides an alternative to increasing sample size for GWAS by composite analysis, unifying two aspects of the LD phenomenon: strength of association and number of surrounding significant SNPs. A genomic convergence approach mapping genetic association signals on expression data, and other biological studies, can then be used for verification of particular genes. The concept is that multiple datasets with results pointing in the same direction is evidence of a true scientific finding. In genomic studies, the most frequent datasets that have been converged are genetic association studies highlighting a candidate gene and the expression of that gene in a biologically relevant tissue.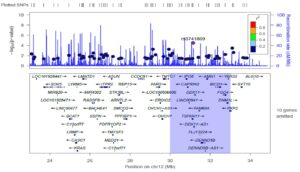
Figure 2 shows the finemapping of the 12p11 locus visualized in LocusZoom
We mapped 410 genes to 30 SLE loci identified using OASIS analysis of two dbGAP GWAS datasets. Using a genomic convergence approach, these genes were investigated for expression in three SLE GEO datasets and a fourth from Cordoba, Spain. This methodology identified DDX11, located at the 12p11 locus, as an SLE susceptibility gene. Interestingly, the 12p11 locus has previously been linked to familial SLE. The DEAD/H box helicase, DDX11, is an RNA helicase involved in genome stability and immunoglobulin diversification. A comprehensive screen of the 12p11 locus (~50K SNPs) in two additional GWAS confirmed DDX11 as a SLE susceptibility gene. Furthermore, it identified DNM1L and its eQTLs to be associated with SLE. DNM1L, involved in mitophagy, is upregulated in SLE. Mitophagy is autophagic removal of dysfunctional mitochondria and is important in immune regulation. Altered mitophagy led to auto-antibody production and lupus nephritis.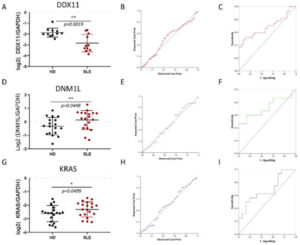
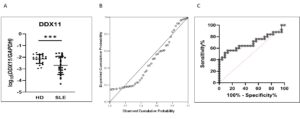
Figure 3 shows the expression of DDX11 is significantly downregulated in SLE
The expression of both DDX11 and DNM1L was confirmed to be significantly altered in SLE by quantitative PCR on SLE monocytes. DDX11 was downregulated whereas DNM1L was upregulated in SLE. Potentially, DDX11 may be employed as a clinical test for SLE in the future after appropriate clinical trials. Convergence of results from several datasets of multiple data types, such as genetic association and expression, provides robust evidence of pathobiological involvement of genes. This study, by utilizing genomic convergence of the 12p11 locus with four European GWAS and four gene expression datasets, followed by eQTL and protein network analysis, identified DDX11 and DNM1L as novel SLE susceptibility genes. This study lays the foundation for understanding the pathogenic involvement of DDX11 and DNM1L in SLE by identifying them using a systems-biology approach, while the 12p11 locus harboring these genes was previously missed by four independent GWAS. Reference: Mohammad Saeed et al. Int. J. Mol. Sci. 2021, 22(14), 7624; https://doi.org/10.3390/ijms22147624Lupus Pathobiology based on Genomics
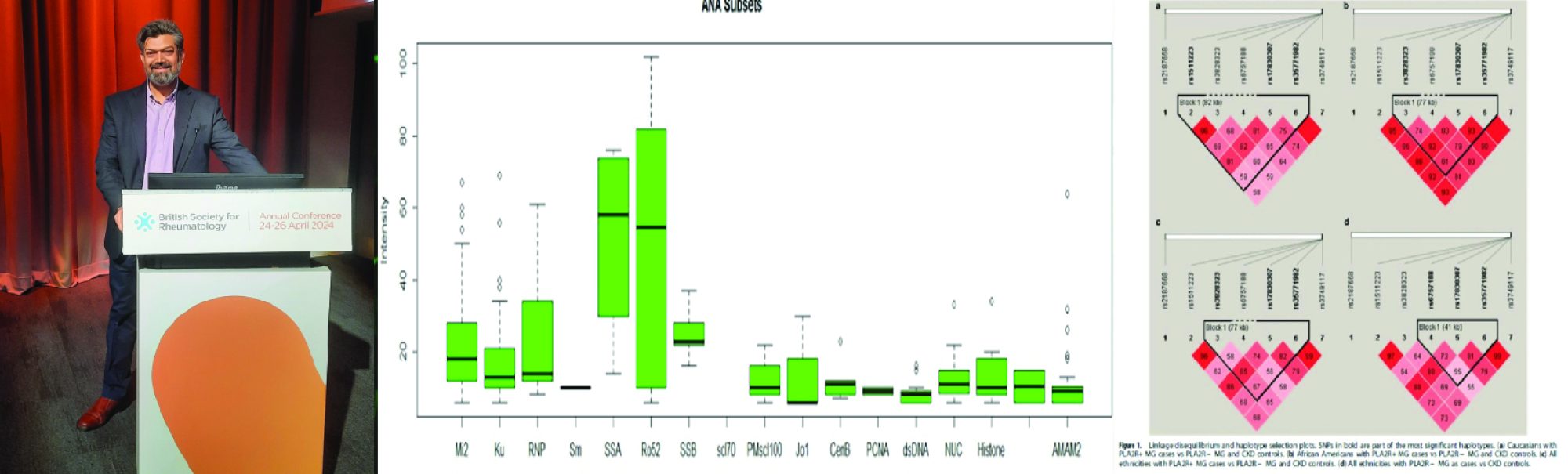
 ‘The challenge will be more robust clinical classification of SLE. As more antibodies are discovered, such as MDA5 and PLA2R, and genetic tests seep into clinical practice, it is likely that SLE classification may change further. Perhaps, SLE clinical definition may incorporate a mandatory presence of nuclear antibodies (ANA and its subsets) and the nuclear-antibody negative SLE may be classified according to the specific antibodies or mutation findings.’…..’On the other hand, expert panels may take a more inclusive approach by subclassifying SLE into more refined phenotypes. Whatever the course SLE classification takes in the future, genetic exploration of this complex disease has not only enhanced its own pathobiology but opened doors to understanding autoimmune disease in general as well.’ Mohammad Saeed, Dec 2017, Immunogenetics.
Lay Summary of the Lupus Review
‘The challenge will be more robust clinical classification of SLE. As more antibodies are discovered, such as MDA5 and PLA2R, and genetic tests seep into clinical practice, it is likely that SLE classification may change further. Perhaps, SLE clinical definition may incorporate a mandatory presence of nuclear antibodies (ANA and its subsets) and the nuclear-antibody negative SLE may be classified according to the specific antibodies or mutation findings.’…..’On the other hand, expert panels may take a more inclusive approach by subclassifying SLE into more refined phenotypes. Whatever the course SLE classification takes in the future, genetic exploration of this complex disease has not only enhanced its own pathobiology but opened doors to understanding autoimmune disease in general as well.’ Mohammad Saeed, Dec 2017, Immunogenetics.
Lay Summary of the Lupus Review
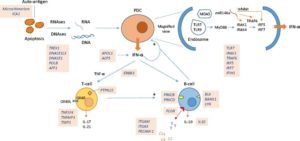
- Lupus is a syndrome rather than a single disease. 330 types of lupus are possible. Therefore, every lupus patient is unique.
- Lupus is initiated with aberrant processing of DNA (and RNA) during apoptosis. This means the clearance system of the body is defective and unable to remove properly the debris of dying cells.
- This nuclear material is picked up by cells of the immune system specifically plasmacytoid dendritic cells (pDCs) and B-cells which react to it (through intricate cellular pathways) by releasing high levels of interferon-alpha.
- These cells may be further activated by viral and bacterial infections or conditions like pregnancy.
- Both pDCs and B-cells present the nuclear material in immunoreactive forms (antigens) to T-cells which coordinate the formation of antibodies by the B-cells.
- The cells and antibodies attack the ‘self’-antigens in tissues such as the kidney and brain.
- This sets up a perpetual cycle of immunologic abnormalities that defines lupus – aberrant apoptosis followed by immune complex formation and deposition.
- Antibodies are often present in lupus but do not necessarily cause lupus. They however do indicate a dysfunctional immune system. SLE can result even without any detectable antibodies.
- SLE treatment has so far largely been guided by clinical experience. With the recent advent of targeted biologic therapies, this paradigm has started to change.
- Molecular analysis has shown the similaries and differences between various SLE drugs. Biologic medications such as Rituximab, Belimumab (both anti-B cell) and the upcoming Anifrolumab (designed against the interferon alpha receptor – anti-pDC and B-cell) are powerful lupus treatment medications. Anifrolumab was shown to be successful treatment for SLE in a 2020 Trial
- Given more than 100 genes now known for lupus and a plethora of antibodies, it is expected that the clinical disease definition of lupus will change as well. The new definition may be more clinically robust.
OASIS
A Novel Method of GWAS Analysis
Mohammad Saeed, MD
Search engine for Oasis in Gene Deserts
Saeed, M. Immunogenetics (2017). Python Code
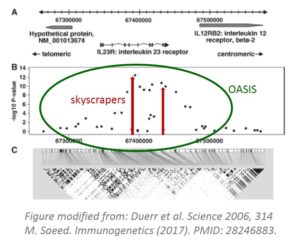
The OASIS concept
Linkage disequilibrium (LD) is the fundamental concept driving genetic association studies. Its effects are clearly seen in this graph. The interesting aspect is that there are several smaller peaks creating a cluster around APOE4, which stands like a skyscraper.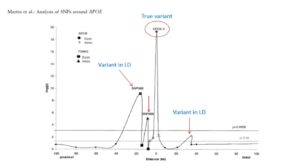
OASIS Software
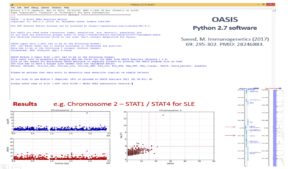 Classically in GWAS, skyscrapers are searched for using varying cutoffs such as Bonferroni and FDR. OASIS looks for LD clusters which are likely more robust evidence of the presence of a disease causing gene.
Classically in GWAS, skyscrapers are searched for using varying cutoffs such as Bonferroni and FDR. OASIS looks for LD clusters which are likely more robust evidence of the presence of a disease causing gene.
OASIS Presentation
The presentation describes the background, concept, methodology and software of OASIS (click 'green' link above).
Post Reference: Saeed, M. Immunogenetics (2017) 69: 295-302. PMID: 28246883.OASIS Application
Using OASIS novel genes and loci for several complex disorders were discovered. These include separate publications on Lupus, Diabetic Nephropathy, Parkinson's Disease and Psoriasis. A novel study identifying DDX11 as a new gene for Lupus is submitted for publication. It shows that a region on chromosome 12p harboring DDX11 was identified using the OASIS methodology and verified by gene expression analysis in monocytes from SLE patients.Fractal Genomics
A Novel Method to Study Evolution of Genes
Mohammad Saeed, MD
Saeed, M. Immunogenetics. Pre-print bioRxiv (2020). Python Code
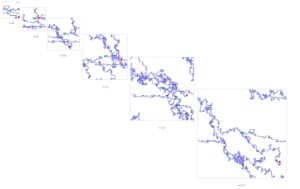 Lay Summary
DNA is a sequence of nucleotides A, T, G and C assumed to be arranged randomly. However, patterns were detected in the order of nucleotides in DNA in 1992. It was not clear why. Few theories were proposed. This paper debunks those theories and proposes a novel explanation - determinsitic chaos (Fractals) as in weaving or knitting with small strings of different colors. This process did not happen once but multiple times throughout evolution. In other words the wheel was invented many times and every time was somewhat different.
Fractal Genomics
The extraordinary complexity of biological systems is generated by repeating patterns coded by recursive processes which appear irregular and random however, are in fact governed by rules of chaos – self-similarity, sensitivity to initial conditions and long-range order over multiple scales of space and time (fractals). Random mutation and natural selection are known to be two forces that have shaped evolution. How these forces led to the development of fractals in DNA sequences is an intriguing question. Fractal Genomics helps to understand how functional genomic sequences developed from short strands of assorted nucleotides.
Long-range correlations (LRC) exist in DNA however, their purpose and mechanism(s) of development is not clearly understood. Earlier thought to occur due to segmental duplication and modified by point mutations Dr. Mohammad Saeed argues that they play minor roles if any in the evolution of natural genomic sequences such as SOD1. His recent study shows that even short range correlations are present in DNA (in small sized sequences from lower organisms) and that genes, e.g. SOD1 did not evolve by patching together sequences from simpler organisms. Instead, SOD1 evolved multiple times over the course of evolution by repeated unique assemblies of oligomer sequences, consequently leading to the development of LRC.
Fractal diagrams of SOD1 gene orthologs can be viewed and downloaded by clicking the link.
Lay Summary
DNA is a sequence of nucleotides A, T, G and C assumed to be arranged randomly. However, patterns were detected in the order of nucleotides in DNA in 1992. It was not clear why. Few theories were proposed. This paper debunks those theories and proposes a novel explanation - determinsitic chaos (Fractals) as in weaving or knitting with small strings of different colors. This process did not happen once but multiple times throughout evolution. In other words the wheel was invented many times and every time was somewhat different.
Fractal Genomics
The extraordinary complexity of biological systems is generated by repeating patterns coded by recursive processes which appear irregular and random however, are in fact governed by rules of chaos – self-similarity, sensitivity to initial conditions and long-range order over multiple scales of space and time (fractals). Random mutation and natural selection are known to be two forces that have shaped evolution. How these forces led to the development of fractals in DNA sequences is an intriguing question. Fractal Genomics helps to understand how functional genomic sequences developed from short strands of assorted nucleotides.
Long-range correlations (LRC) exist in DNA however, their purpose and mechanism(s) of development is not clearly understood. Earlier thought to occur due to segmental duplication and modified by point mutations Dr. Mohammad Saeed argues that they play minor roles if any in the evolution of natural genomic sequences such as SOD1. His recent study shows that even short range correlations are present in DNA (in small sized sequences from lower organisms) and that genes, e.g. SOD1 did not evolve by patching together sequences from simpler organisms. Instead, SOD1 evolved multiple times over the course of evolution by repeated unique assemblies of oligomer sequences, consequently leading to the development of LRC.
Fractal diagrams of SOD1 gene orthologs can be viewed and downloaded by clicking the link.
SOD1 Evolution in multiple clades
Gene Fractals Software
The code can be downloaded from GitHub under the GNU 3.0 Public License.
This is the Python 2.7.9 code for the software, GeneFractals. It performs the following analysis on genetic sequences:
1. k-mer analysis with comparison between genetic sequences (Homolog.txt and Folder Stats)
2. Relative Dispersion Analysis (RDA.txt)
3. Detrended Fluctuation Analysis (DFA.txt, Folder DFA)
4. Fractal Gene Diagrams (Folder Diag)
It requires the following files:
1. FASTA files of genetic sequences to be investigated (without headers)
2. Compiled dfa.exe application along with a dfa.bat file containing the following code:
dfa 0dfa.txt
3. A text file names FILELIST.txt containing the list of filenames of FASTA sequences